Previous
Previous
|
Students
Nhat Vu, Elec & Comp Engineering
Wes Smith, Media Arts & Tech
Dan Overholt, Media Arts & Tech
|
Faculty
Advisors
B.S. Manjunath, Elec & Comp Engineering
Steve Fisher, Molecular Cellular Developmental Biology
Stephen Pope, Media Arts & Technology
George Legrady, Media Arts & Technology
|
Next
 |
| |
Abstract
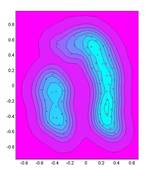
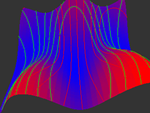
In many applications ranging from biological image processing to interactive audiovisual composition and performance, high dimensional data or features are generated and analyzed to make predictions and decisions. However, to gain visual insight into the behavior of the data or to interactively navigate the feature space, the original data must be reduced down to two or three dimensions for visualization. Often dimensionality reduction is a tradeoff between how much information is lost versus the simplicity of visual representations. Hence the reduced data may not wholly depict the behavior of the original dataset. In this project, we investigate a widely used data reduction technique known as Principal Component Analysis (PCA). PCA projects the original high dimensional data onto a smaller subset of dimensions that capture most of the variations or energy in the data. However, PCA makes certain assumptions about the data that are not always true in practical settings. Consequently using only the principal axes as a guide when visualizing or synthesizing new data points may yield misleading behaviors. To enhance the visualizations, we use kernel PCA (kPCA) to calculate variation isocontours and probability maps, which can characterize the behavior of the data more accurately. Having this common data visualization framework, we seek to analyze datasets from our respective research components.
The project consists of two components: Bio-image processing in which we are analyzing biological images to study retinal detachment, and an audiovisual performance based on the same tools and techniques so as to formulate artistic expression from the science. For the first component, our dataset contains a collection of images taken at various times during detachment experiments. The scientific goal is to model the underlying processes producing these images and to visually simulate retinal restructuring during detachment. As an example, given an image of a tissue sample after three days of detachment, we want to produce an image sequence that extrapolates the sample backward in time to day one or forward in time to day seven. The underlying process parameters must evolve in time to produce convincing images interpolating various stages of the experiment. The data visualization can provide us with intuition on how the model parameters change during detachment and consequently may reveal underlying biological processes. For the audiovisual composition and performance component, we are developing custom input devices with high bandwidth sensors that are designed to control an interactive performance. The theme of the performance will be looking at the relationships between biological imagery and the corresponding measurement devices, mapping variations in the images and user input to corresponding noise and pure sounds, and the degree of coherence between light and sound components of the experience. We are using the kPCA framework to explore methods of input data reduction and feature extraction so that precise, high-level information about user interaction can be passed on to the audiovisual processing system. This framework allows us to create mappings from sensor input to parameter control with a high degree of sensitivity toward expressive user interactions in both the audio and visual domains.
|
|

|
|
|
|